Mission statement
The mission of my lab is to understand, at systems level, how the extracellular microenvironment modulates cell shapes and behaviours through the transduction of proteomic information from the extracellular space through the plasma membrane into the cell nucleus.
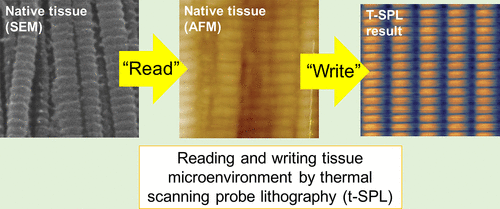
My lab has shown that the differentiation of adult stem cells and the migration of cancer cells can be controlled by the surface topography of cell culture substrates (Tong et al, 2012, Lam et al, 2020). In particular, substrate topographies that mimic those of the native tissue microenvironment can differentiate mesenchymal stem cells, even in the absence of any biochemical factors (Tang et al, 2019). Our working hypothesis is that it is the precisely controlled physical microenvironments such as shape and texture provided by the extracellular matrix (ECM), not the presence of biochemical ligands, that functionally define a tissue microenvironment (reviewed in Tang et al, 2020). This explains the paradoxical observation that the functions of a tissue microenvironment, such as in decellularised organs (Shen et al, 2020), still persist even after it is biologically denatured. To understand the biochemical principle of this topographical information, our lab is currently mapping the abundance of 600 ECM components in normal and cancer tissues in humans. We discovered that each tissue expresses a unique signature of ECM proteins, which is altered in cancer (manuscript in preparation). This information will guide the design of future ECM array experiments that are more physiologically relevant than the current attempts.
In parallel, our lab is interested in how the cell nucleus, a repository of the genome and a major location of gene expression regulation, is organised into functional compartments without any membrane (unlike cyto-
plasmic organelles). Using the nucleolus as a model system, we have studied the flux of proteins into and out of nuclear compartments by using proteomics, isotope incorporation and live-cell microscopy (Andersen and
Lam et al, 2005, Lam et al, 2007, Lam et al, 2010, Kar et al, 2011, Liang et al, 2012). We have also discovered a class of microRNAs that is sequestered in the nucleolus and released into the cytoplasm after viral infection (Li et al, 2015 and manuscript in preparation). Many of these data suggest that macromolecules within the nucleus is self-organised into “territories” that are maintained by molecular interactions between some of the key architectural factors. This is compatible with the recent theory of liquid-liquid phase separation (LLPS) as the organisation force of the cell structure. To test this theory, we argue that changes in the chemical environment that interferes with the phase separations of nuclear factors will alter gene expression. For example, we have recently shown that divalent cations in the tissue microenvironment, such as magnesium ions, alone is sufficient to cause cell type-specific changes in gene expression and epigenetic modifications (Wei et al, 2021) and that the assembly of nuclear structures can be recapitulated in a cell-free system by using non-coding RNA in a molecularly crowded microenvironment (manuscript in preparation).
Taken together, my research is motivated by the understanding of how the physiochemical environment influences the assembly of intracellular structures and the coordination of tissue functions, and how the dysregulation of this “haptic” information contributes to pathology. We seek to highlight the importance of physical conditions, such as molecular crowding, ionic strength, and surface topographies, on the top of the more traditional, gene-centric approach in biology.
Besides, through an innovative application of molecular cell biology techniques, from quantitative proteomics to live-cell imaging, we have played a central role in many mulitidisciplinary projects. These ventures have taken us to many exciting territories in biology, from tissue regeneration to chemical biology, from drug development to molecular psychology. Please click here to see our major research interests.